CHKBIO’s Novel cronavirus(COVID-19) RT-PCR detection kit(Lyophilized) are designed at the conserved region of RdRP gene, N gene of COVID-19 virus, the kit can detect all the mutated strains found globally till now.
Our R&D team already checked all the mutation sites of the new mutated strains including Alpha GRY(B1.1.7+Q.*, first detected in the UK), Beta GH/501Y.V2(B.1.351+B.1.351.2+B.1.351.3, first detected in South Africa), Gamma GR/501Y.V3(P.1+P.1.*, first detected in Brizal/Japan), Delta GK(B.1.617.2+AY.*, first detected in India), Lambda GR/452Q.V1(C.37+C.37.1, first detected in Peru), Mu GH (B.1.621+B.1.621.1, first detected in Colombia), GH/490R (B.1.640+B.1.640.*, first detected in Congo/France), Omicron GRA(B.1.1.529+BA.*+XBB.1.5, first detected in Botswana/Hong Kong/South Africa), JN.1. And all these mutation sites are not at our kit’s detecting target sites. So our kit will not be affected by the mutation and the kit can detect all above mutation strains correctly.
Compared with the reference genome WIV04 (sequence number EPI_ISL_402124), the most mutation sites happened on the S gene and ORF1ab gene of COVID-19 virus. Our kits are detecting RdRp (it is a part of ORF1ab gene, Among Nucleotide area:15300-16000 ), N (Among Nucleotide area:28300-28900 ).
The major mutation sites of the new South Africa 501Y.V2 mutation strain in is in S gene. Our COVID-19 RT-PCR kit detecting sites are in RdRP gene and N gene, so it will not affect our detection.
Following the emergence of new SARS-CoV-2 variants, we assessed the impact of the mutations. We can confirm the accurate detection of all these mutation variants.
.
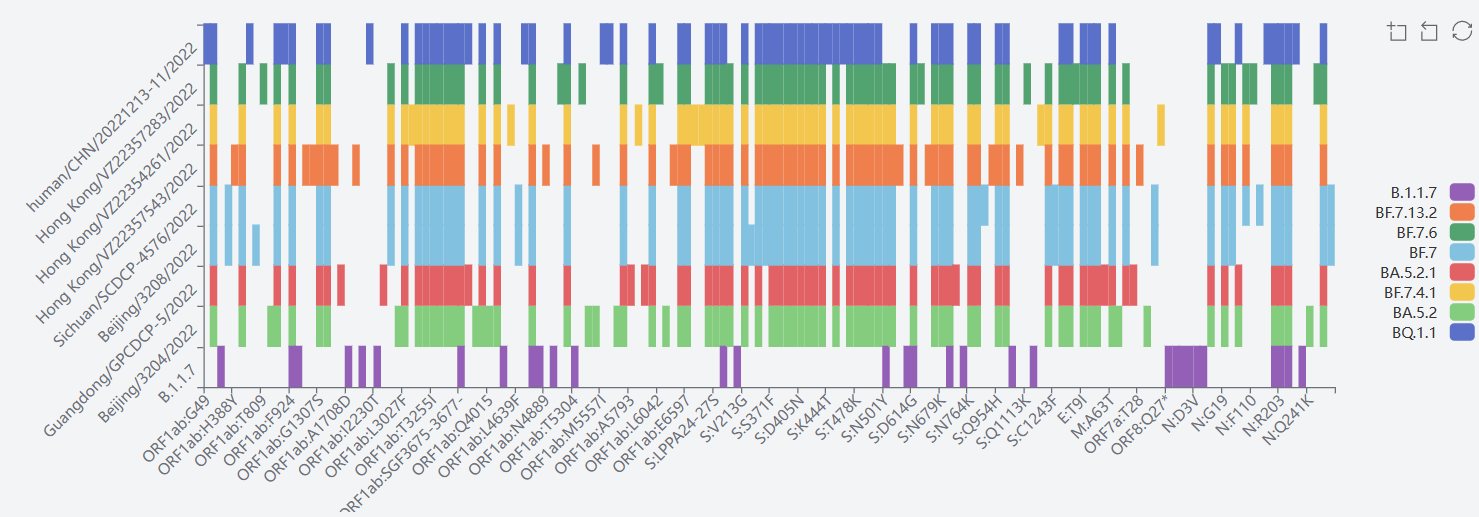
1) Alpha GRY(B1.1.7+Q.*)
The mutation sites of UK mutation strain B.1.1.7
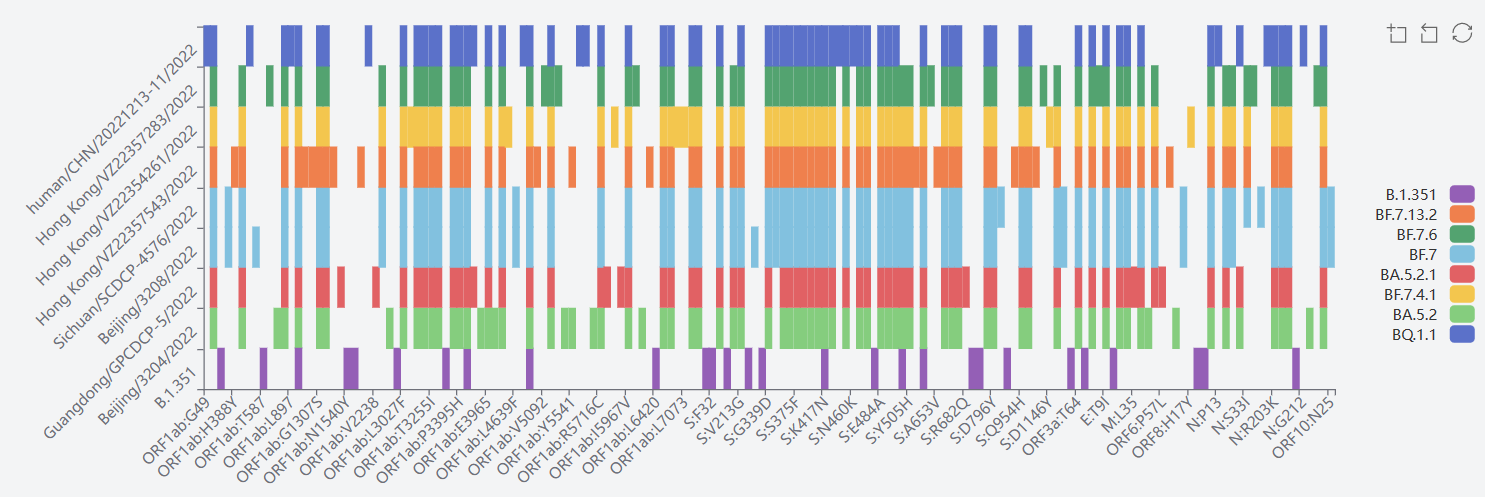
2) Beta GH/501Y.V2(B.1.351+B.1.351.2+B.1.351.3)
The mutation sites of B.1.351
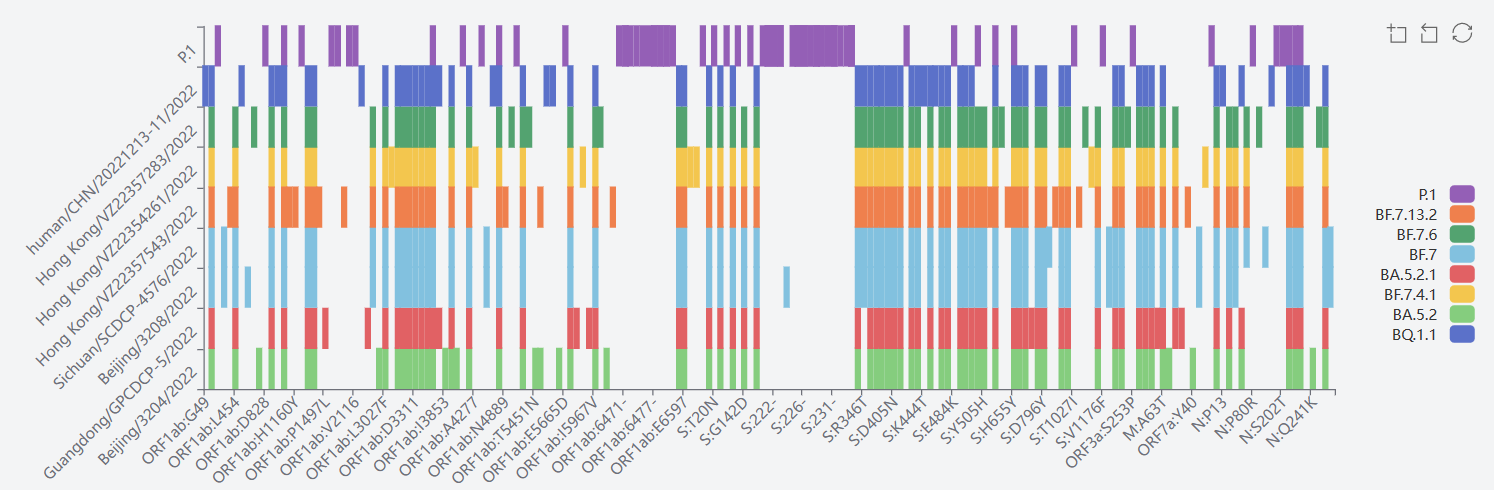
3) Gamma GR/501Y.V3(P.1+P.1.*)
The mutation sites of P.1
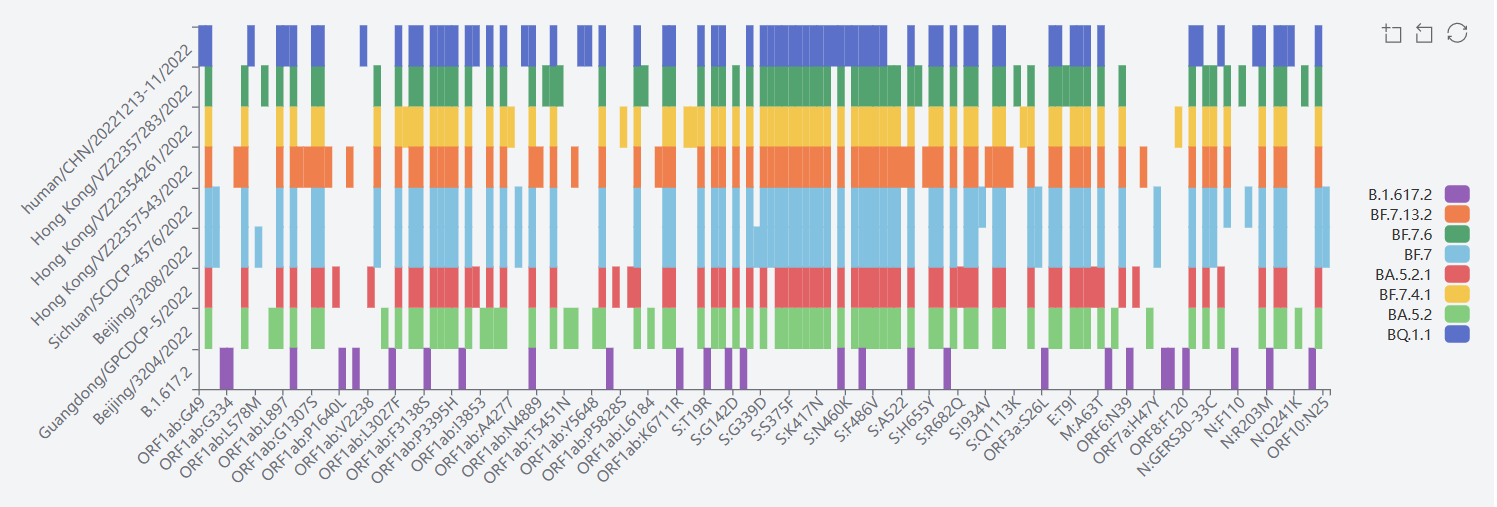
4) Delta GK(B.1.617.2+AY.*)
The mutation sites of Delta (B.1.351)
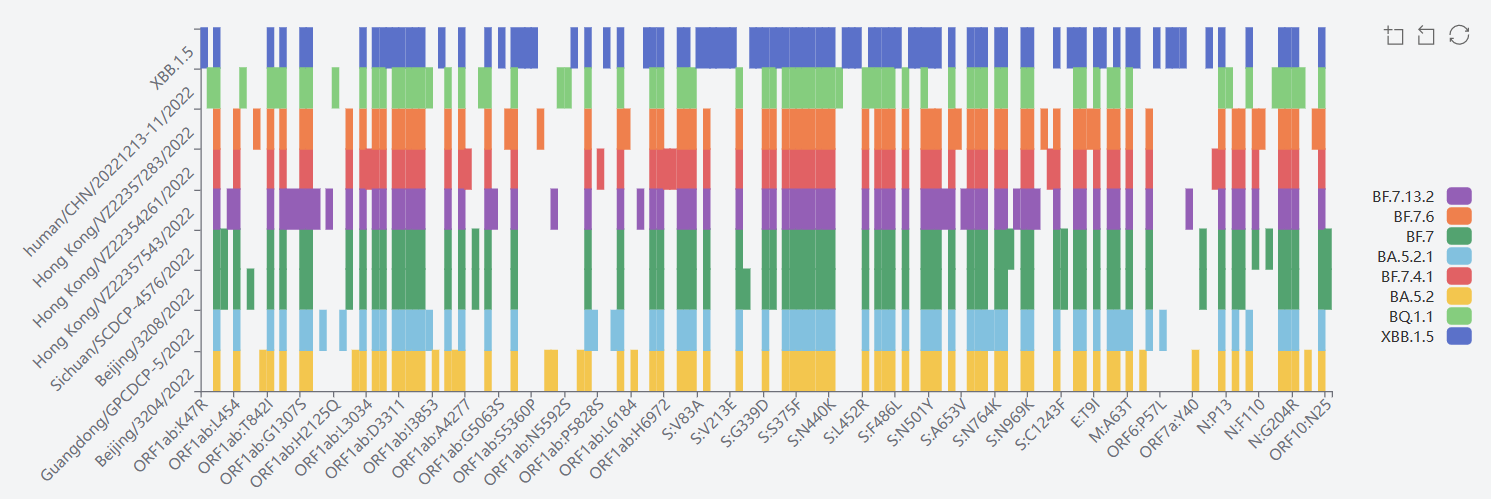
5) Omicron GRA(B.1.1.529+BA.*+XBB.1.5)
The mutation sites of XBB.1.5
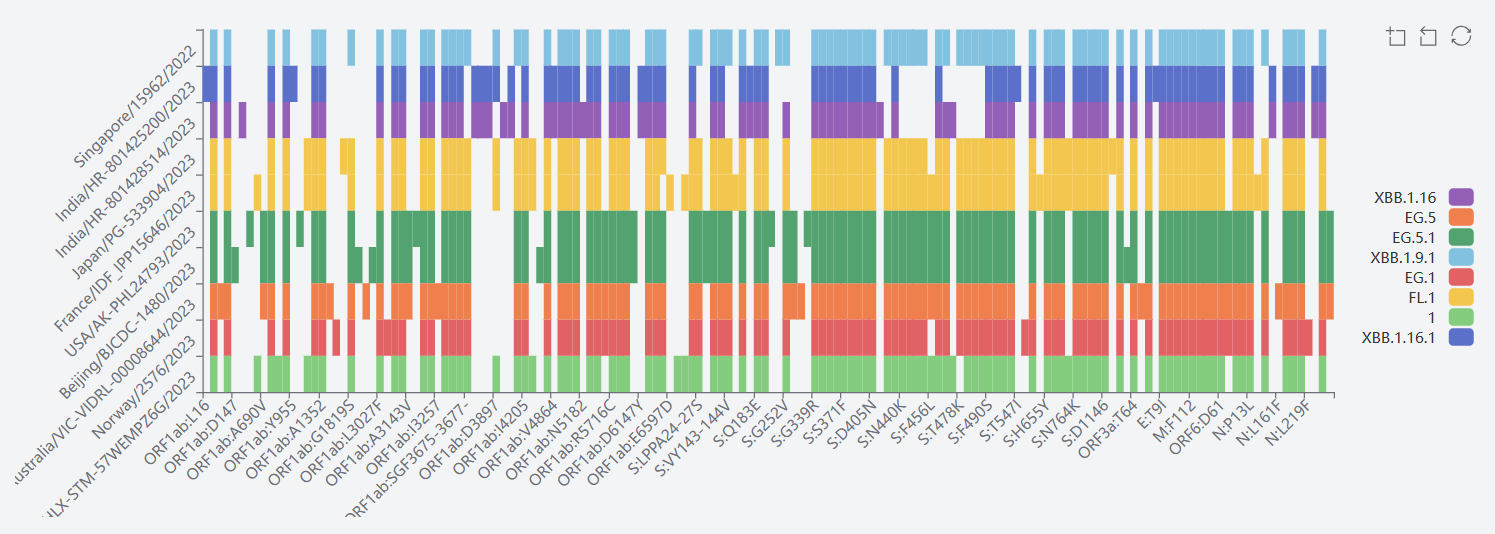
6) JN.1
The mutation sites of JN.1
Post time: Jan-02-2024

 中文
中文